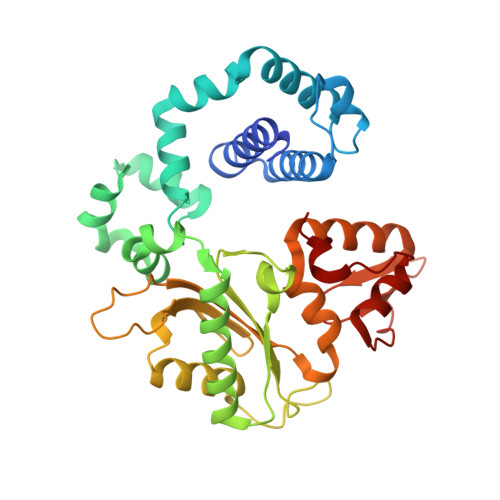
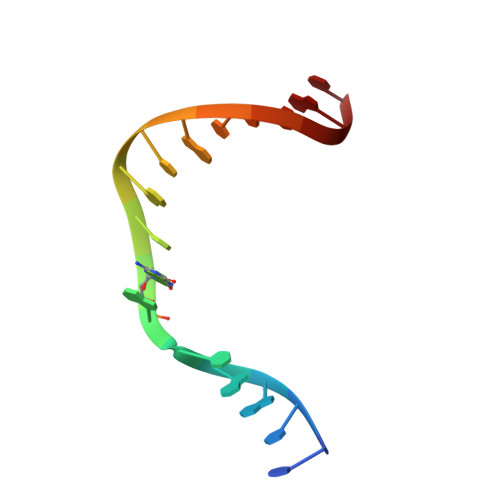
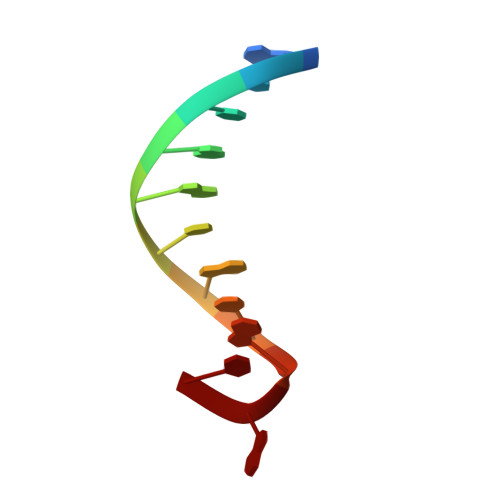
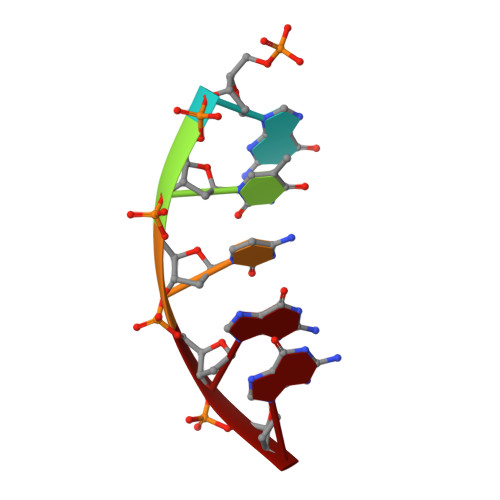
Structure of a DNA polymerase abortive complex with the 8OG:dA base pair at the primer terminus.
Batra, V.K., Wilson, S.H.(2020) Commun Biol 3: 348-348
- PubMed: 32620932
- DOI: https://doi.org/10.1038/s42003-020-1080-4
- Primary Citation of Related Structures:
6W2M - PubMed Abstract:
Adenine frequently pairs with the Hoogsteen edge of an oxidized guanine base (8OG) causing G to T transversions. The (syn)8OG:dA base pair is indistinguishable from the cognant base pair and can be extended by DNA polymerases with reduced efficiency. To examine the structural basis of this reduced efficiency, we sought to obtain the structure of the "product" complex of DNA polymerase (pol) β with the (syn)8OG:dA base pair at the primer terminus by soaking the binary complex crystals with a hydrolysable dCTP analogue complementary to the template base G. Crystallographic refinement of the structure revealed that the adenine of the (syn)8OG:dA base pair had been expelled from the primer terminus and a dCMP was inserted opposite 8OG in a reverse orientation; another uninserted molecule of the analogue was bound to the templating base G. This leads to an abortive complex that could form the basis of oxidatively-induced pol β stalling.
Organizational Affiliation:
Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, 111 T.W. Alexander Drive, Research Triangle Park, NC, 27709, USA.